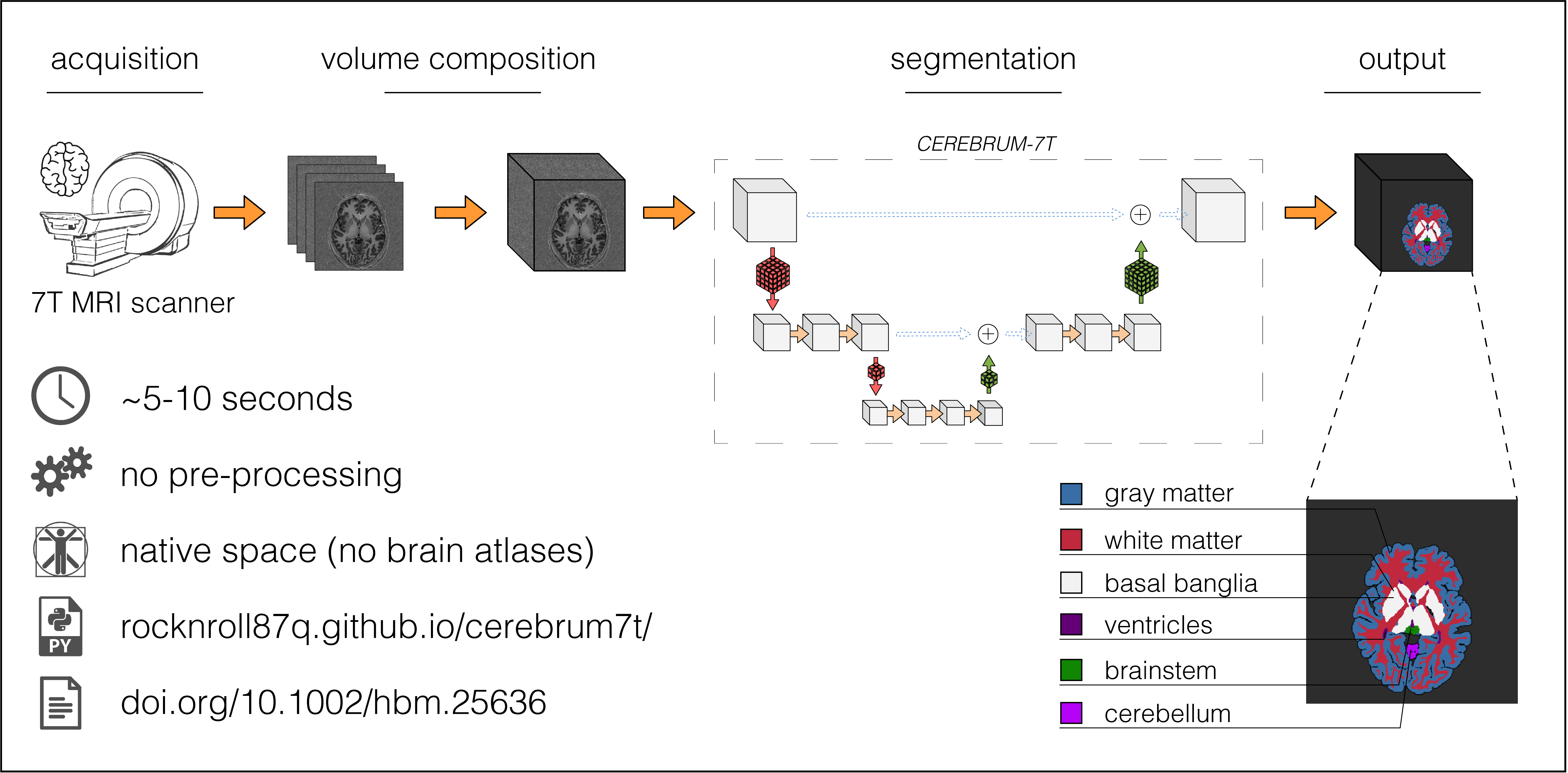
Abstract
Ultra high-field MRI enables sub-millimetre resolution imaging of the human brain, allowing the study of functional circuits of cortical layers at the meso-scale. An essential step in many functional and structural neuroimaging studies is segmentation, the operation of partitioning the MR images in anatomical structures. Despite recent efforts in brain imaging analysis, the literature lacks in accurate and fast methods for segmenting 7 Tesla (7T) brain MRI. We here present CEREBRUM-7T, an optimised end-to-end Convolutional Neural Network (CNN), that allows fully automatic segmentation of a whole 7T T1w MRI brain volume at once, without partitioning the volume, preprocessing, nor aligning it to an atlas. The trained model is able to produce accurate multi-structure segmentation masks on six different classes plus background in only a few seconds. The experimental part, a combination of objective numerical evaluations and subjective analysis, confirms that the proposed solution outperforms the training labels it was trained on, and is suitable for neuroimaging studies, such as layer fMRI studies. Taking advantage of a fine-tuning operation on a reduced set of volumes, we also show how it is possible to effectively apply CEREBRUM-7T to different sites data. Furthermore, we release the code, 7T data, and other materials, including the training labels and the Turing test.
Results
Below, reconstructed meshes of GM, WM, basal ganglia, ventricles, brain stem, and cerebellum of a testing volume, obtained with CEREBRUM7T on sub-013_ses-001 (mesh created with BrainVoyager). A light smoothing operation is performed (50 iterations) - no manual corrections.
In the following pages, there are results of our method with data from three different sites and settings:
- Pag. 1 - training and testing on Glasgow data and
- Pag. 2 - fine tuning experiment on AHEAD data.
- We also push the limit of our method on a very challenging scenario at - Pag. 3.
Pros and Cons
| Timing | Very fast inference: ~ 5/10 sec. |
Training needs ~ 24 hours |
| Hardware | Inference easily done on CPU | Needs 4 GPUs for training |
| Training labels | Does not need accurate labels | Does not overcome systematic errors |
| Visual outcome | Very clean and smooth segmentation | Does not recover from "burn" artefacts |
| New dataset | Works very well but only if... → | Needs fine-tuning |
Usage
Visit the relative page to learn how to use CEREBRUM-7T from source code, docker, or singularity.
Data
Visit the relative page for all the information needed about the data.
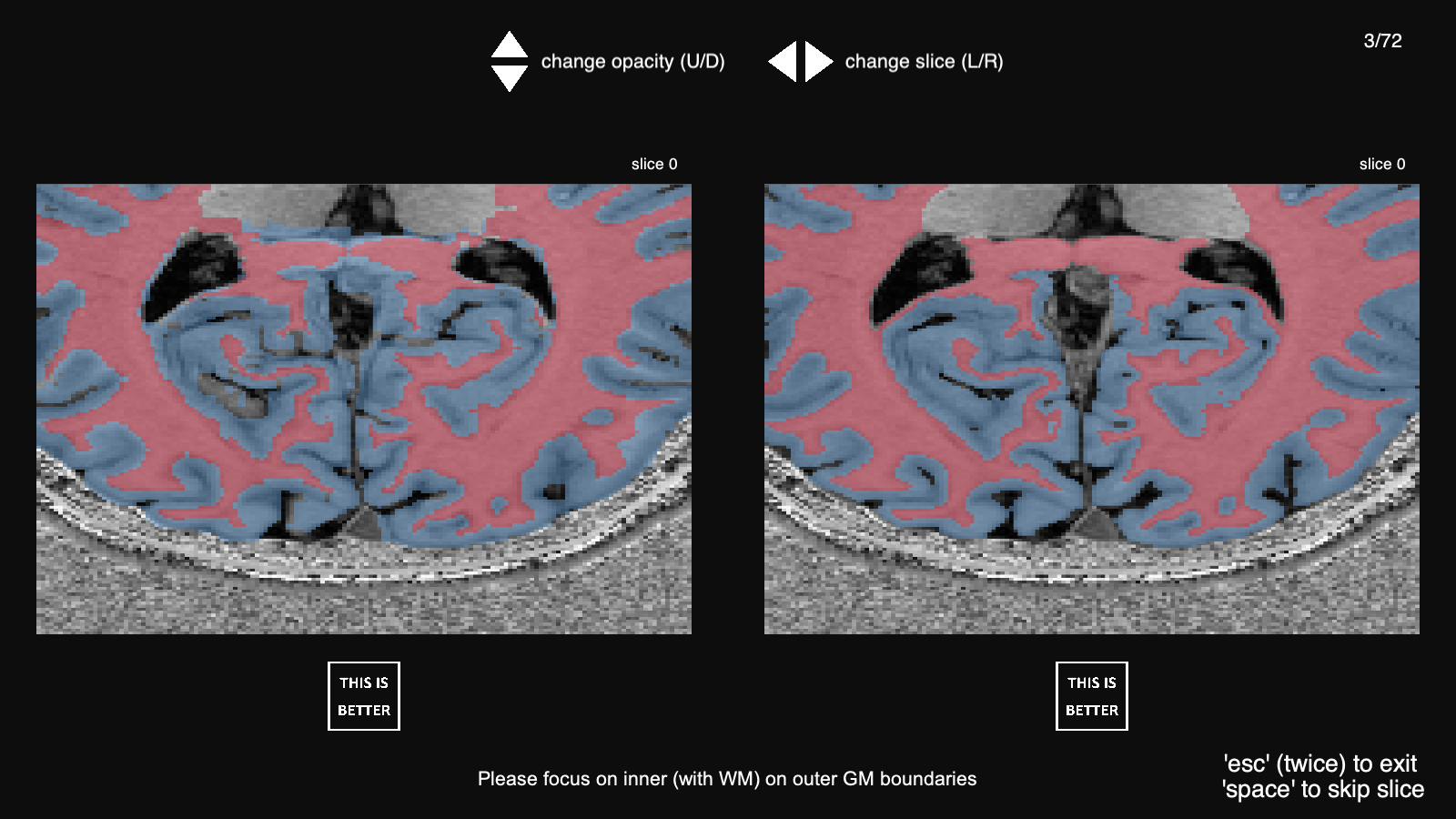
Turing test
As we discussed in the paper, we designed and implemented a PsychoPy (Peirce et al., 2019) test for neuroscientists to assess the segmentation quality of CEREBRUM-7T comparing our model with manual segmentation and with the GT. In the figure below, we show the interface we presented the participants with.
The source code to replicate the test is made available here.
Citation
Svanera, M., Benini, S., Bontempi, D., & Muckli, L. (2021). CEREBRUM-7T: Fast and Fully Volumetric Brain Segmentation of 7 Tesla MR Volumes. Human Brain Mapping, 1– 18. doi.org/10.1002/hbm.25636
@article{SvaneraHBM21Cerebrum7T,
author = {Svanera, Michele and Benini, Sergio and Bontempi, Dennis and Muckli, Lars},
title = {CEREBRUM-7T: Fast and Fully Volumetric Brain Segmentation of 7 Tesla MR Volumes},
journal = {Human Brain Mapping},
volume = {n/a},
number = {n/a},
pages = {},
keywords = {3D image analysis, brain MRI segmentation, convolutional neural networks, weakly supervised learning},
doi = {https://doi.org/10.1002/hbm.25636},
url = {https://onlinelibrary.wiley.com/doi/abs/10.1002/hbm.25636},
eprint = {https://onlinelibrary.wiley.com/doi/pdf/10.1002/hbm.25636},
}
Acknowledgments
This project has received funding from the European Union Horizon 2020 Framework Programme for Research and Innovation under the Specific Grant Agreement No. 785907 and 945539 (Human Brain Project SGA2 and SGA3).