AHEAD: a fine tuning experiment
In this experiment, we accomplish very good results using only 20 volumes to fine tuning the model (trained on Glasgow data) on AHEAD data (link).
The labels used for training were those described in the paper, while the labels for fine tunings derive from FreeSurfer v7.
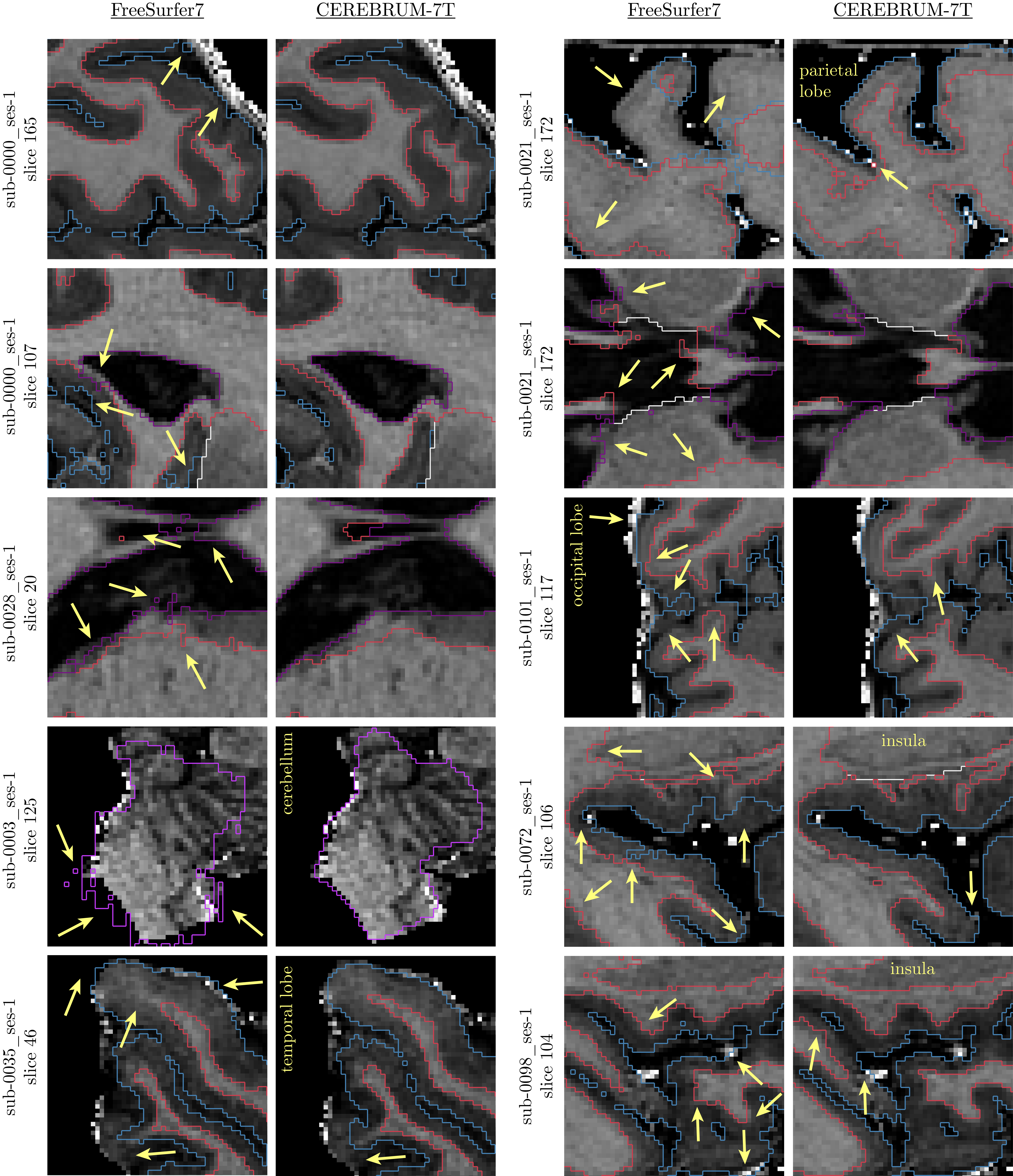
Comparisons that you can see below are made against FreeSurfer v7 on the testing set.
As it is possible to see, FreeSurfer v7, which has been improved for UHF data, is able to segment very well multiple areas (ex. GM/WM boundary), but the inhomogeneity of the scan affects its ability to correctly select all the areas (ex. parietal and temporal lobes). It produces some “holes” in the segmentations. In addition, our method produces much smoother results.
For further inspections, yon can download the segmentation masks for both methods here. The segmented classes and the color code used below are:
| Class ID | Substructure/Tissue | Color |
|---|---|---|
| 0 | Background | Transparent |
| 1 | Grey matter | Light green |
| 2 | Basal ganglia | Dark green |
| 3 | White matter | Red |
| 4 | Ventricles | Blue |
| 5 | Cerebellum | Yellow |
| 6 | Brainstem | Pink |
Results
Result inspection on AHEAD (testing) data. Two sets of columns are shown: FreeSurfer on the left and CEREBRUM-7T on the right. Yellow arrows point to errors. Contour color code is the same used in Figure 2 and Figure 3 for different structures. Please notice that perfectly contiguous structures show overlapping boundaries (so that only one is visible). For example, the inner boundary of GM corresponds to the outer boundary of WM (and only one is displayed).
| Subj ID | ||
| sub_0014 (WM only) | 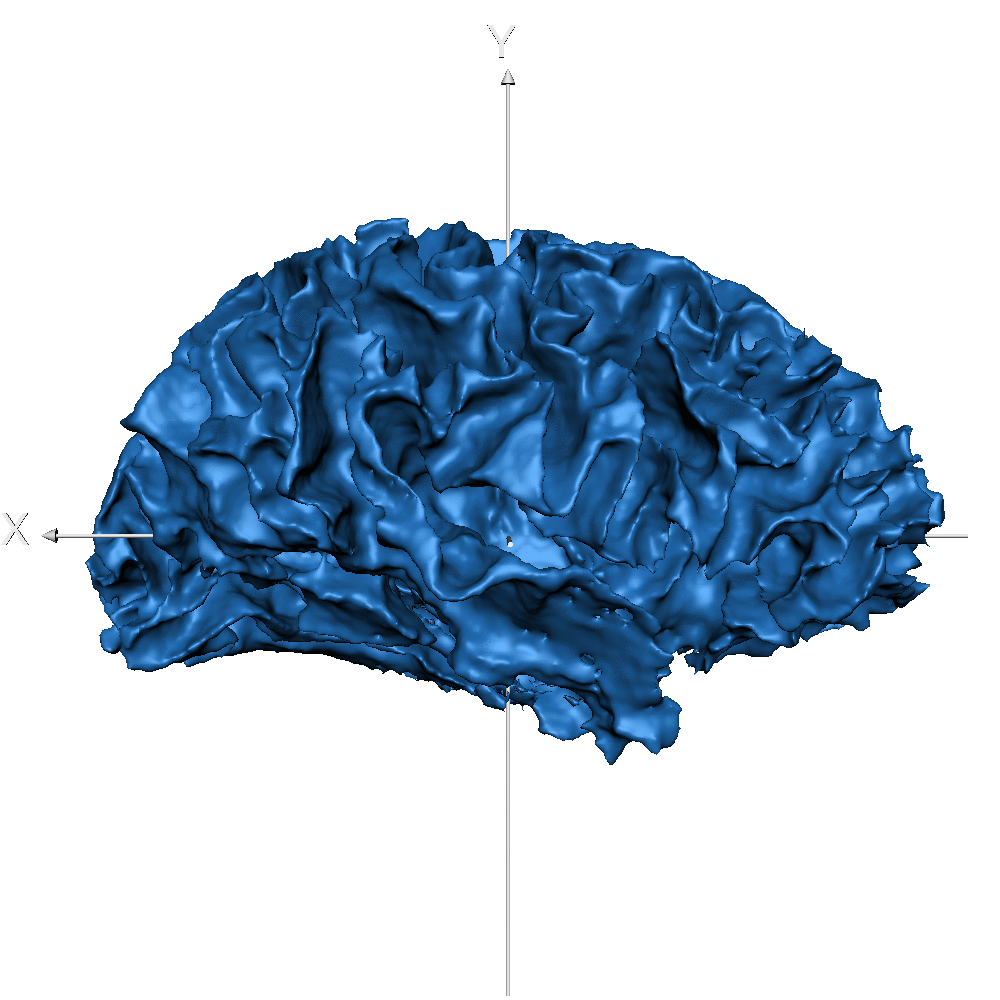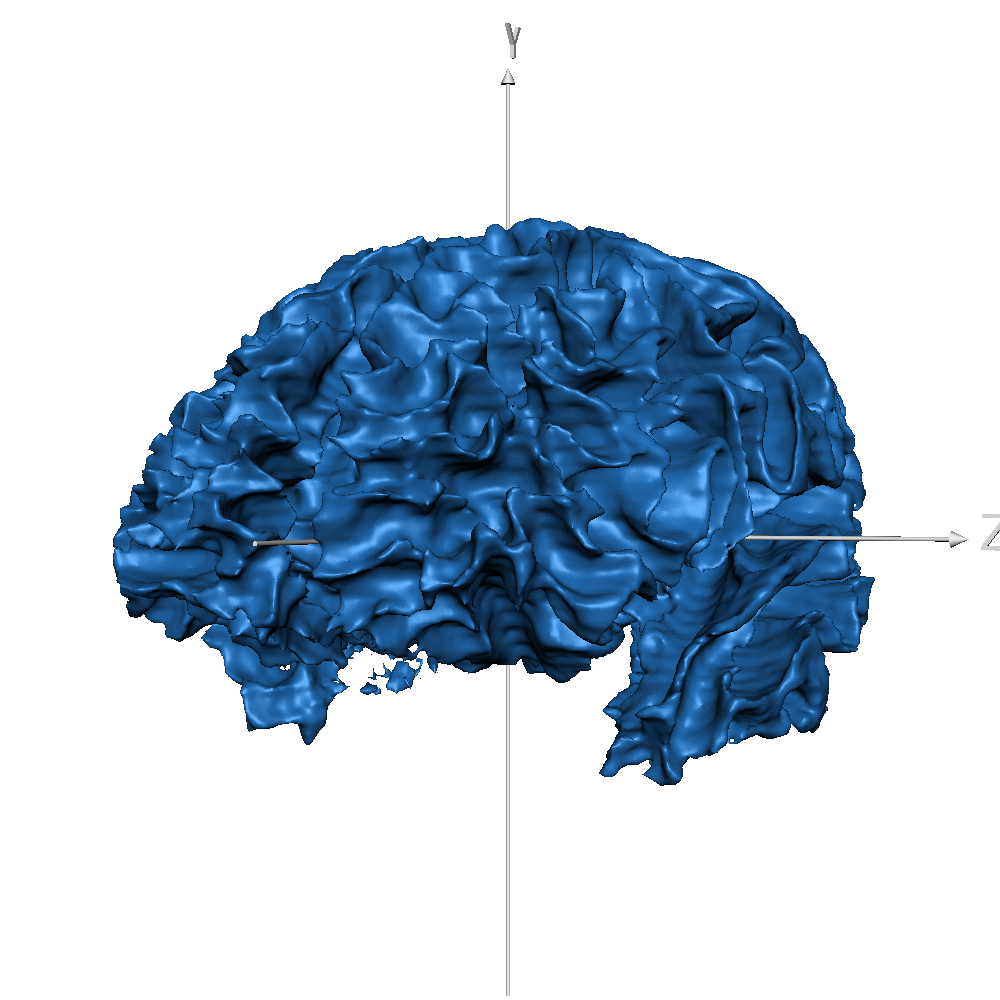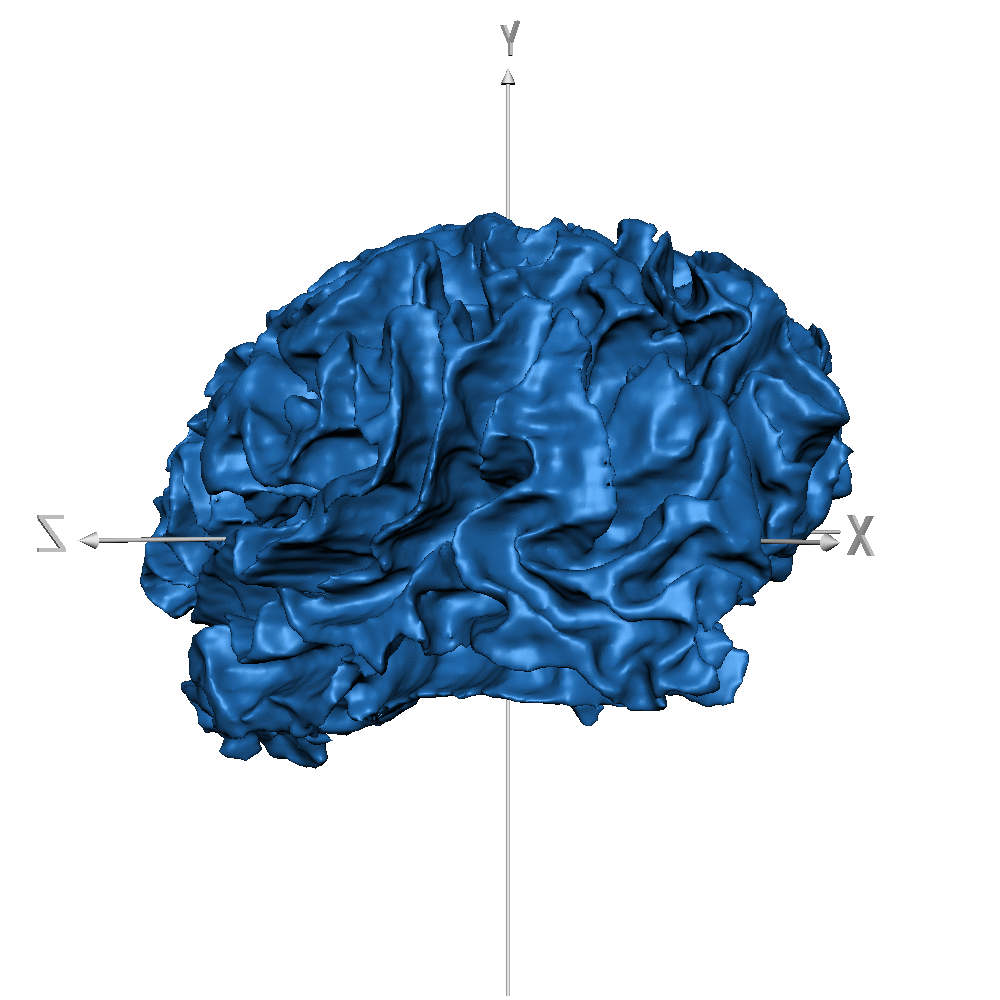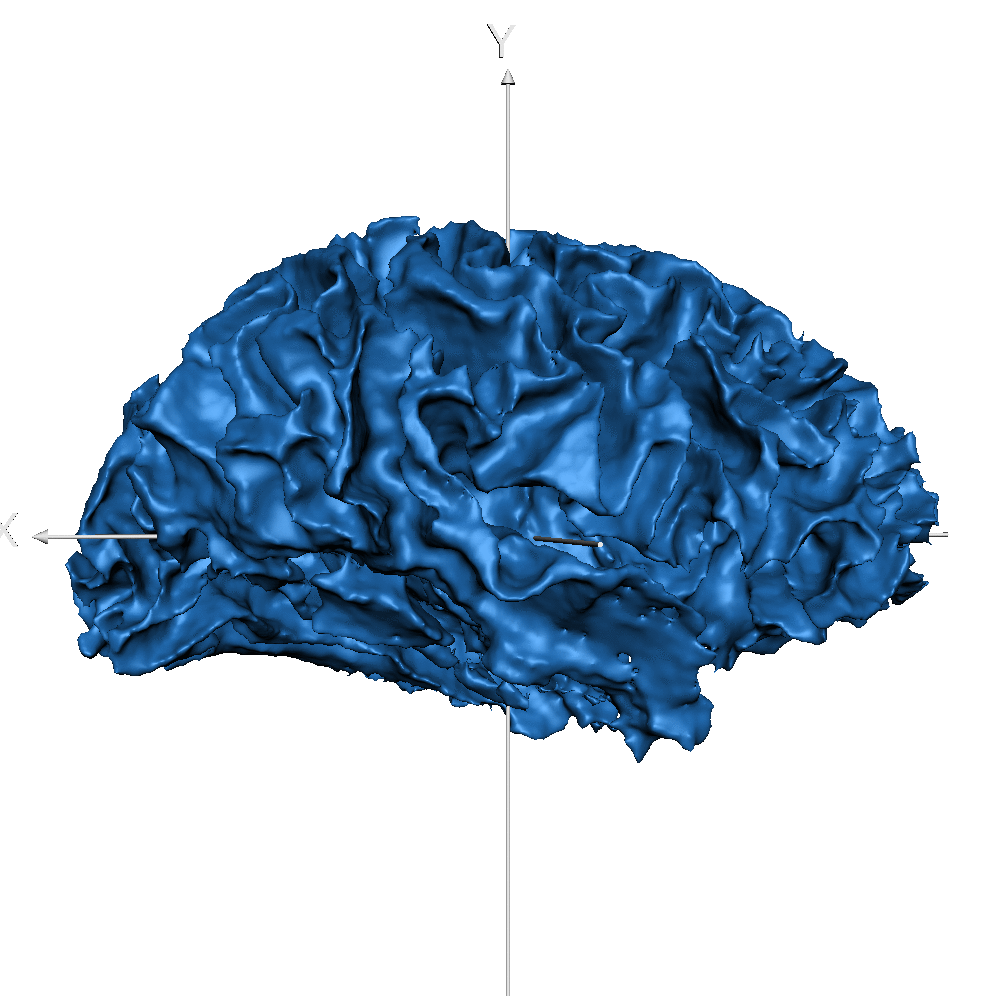 |
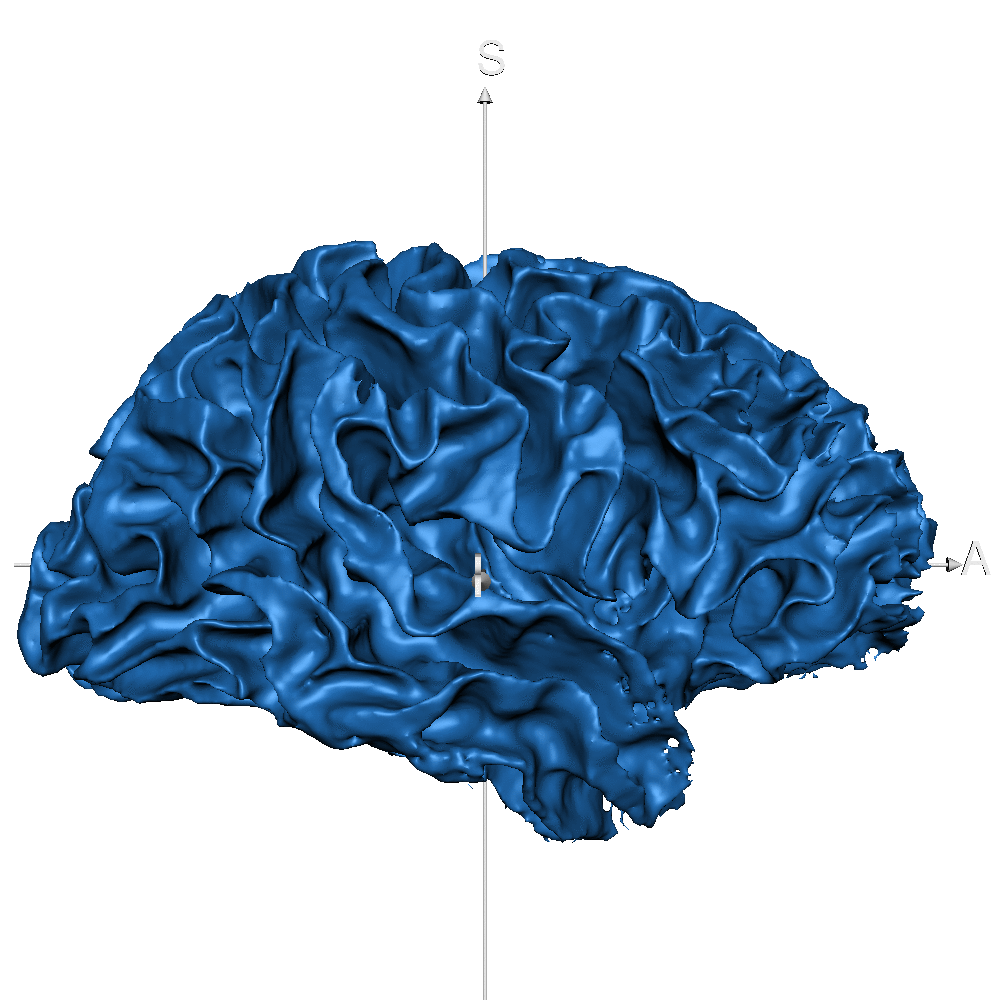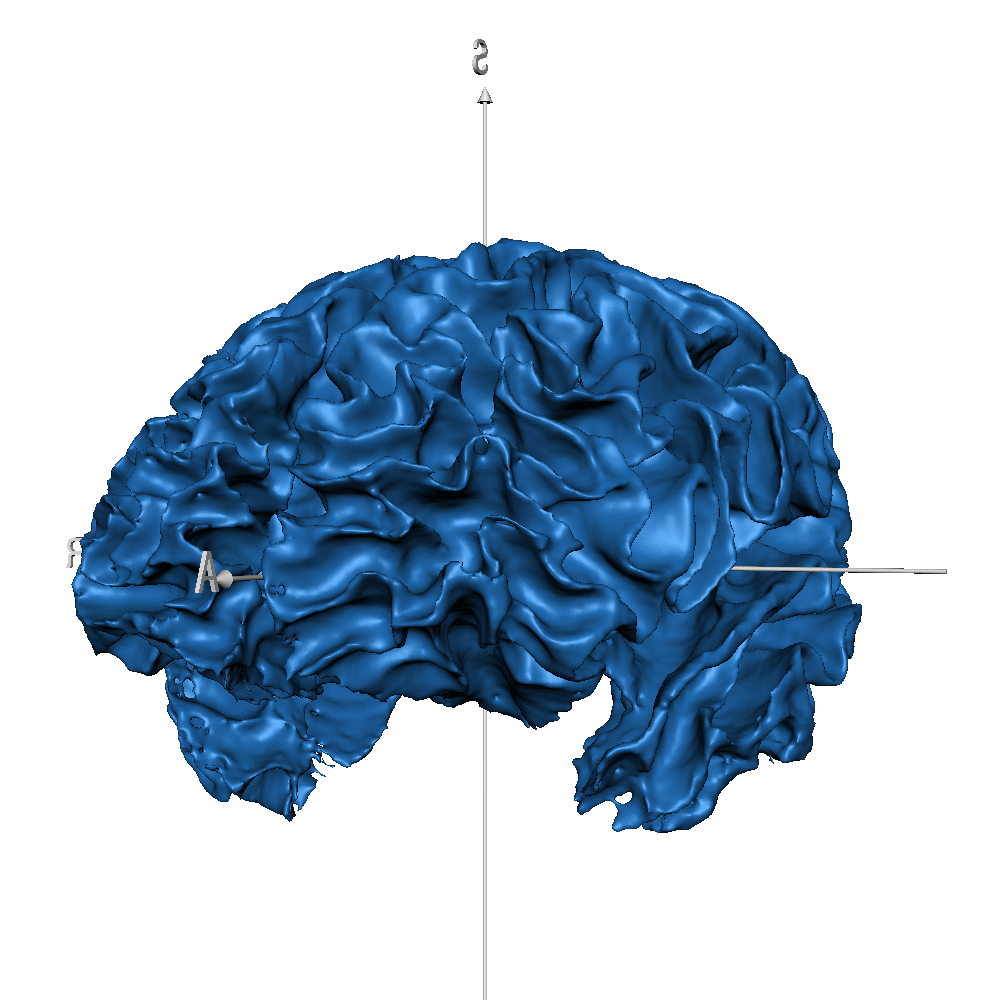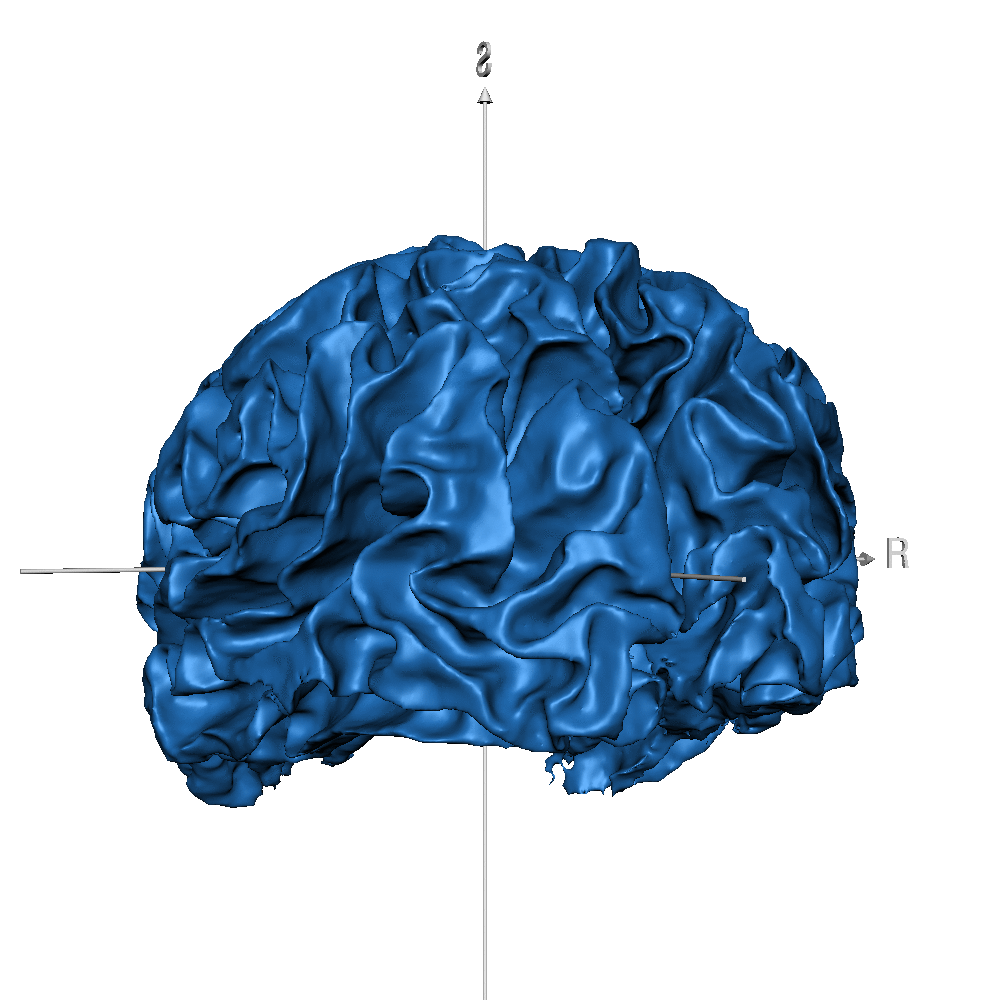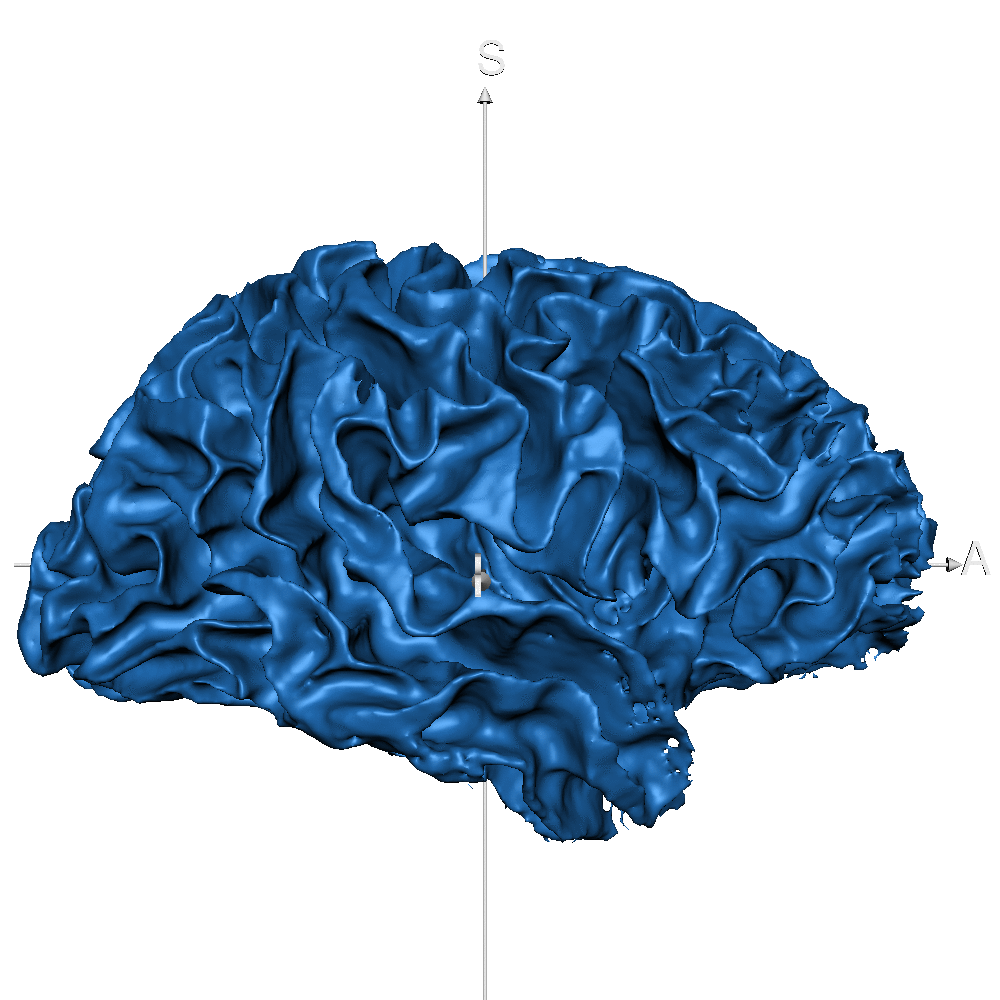 |
| Subj ID | ||
| sub_0000 |  |
 |
| Subj ID | ||
| sub_0014 |  |
 |
| Subj ID | ||
| sub_0025 |  |
 |
| Subj ID | ||
| sub_0035 |  |
 |
| Subj ID | ||
| sub_0011 |  |
 |
| Subj ID | ||
| sub_0037 |  |
 |
| Subj ID | ||
| sub_0038 |  |
 |
| Subj ID | ||
| sub_0040 |  |
 |
| Subj ID | ||
| sub_0068 |  |
 |
| Subj ID | ||
| sub_0093 |  |
 |